GSoC Week 5
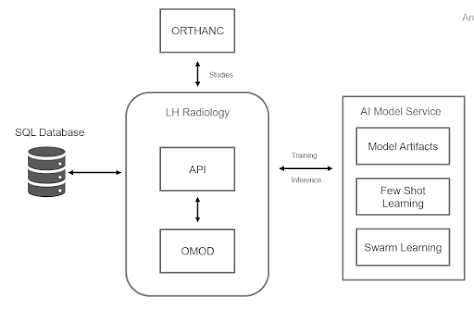
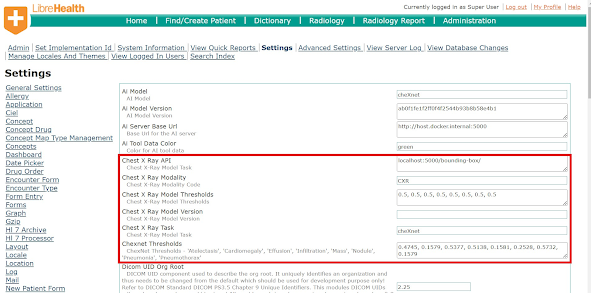
From LH-RIS Viewer to AI Model Service Last week, I primarily focused on coding and making updates to the LH-RIS (Libre Health Radiology Information System) viewer. This week, my work shifted towards the LH AI Model Service. The current architecture of the Flask REST-API within the AI Model Service was unfamiliar to me. It was built in an object-oriented style, which seemed complex compared to the simpler implementations I've encountered in the past. As such, I took my time to understand the system's components before implementation. Implementing the Model Selection API Once I grasped the system, I dived into implementing the Model Selection API. We introduced a few methods on the viewer side that now transmit Dicom tags information and the new model configuration settings, input by the admin in the administration settings. This information is crucial for the Model Selection API, as it determines which model to invoke on the fly. The concept behind the Model Selection API is st